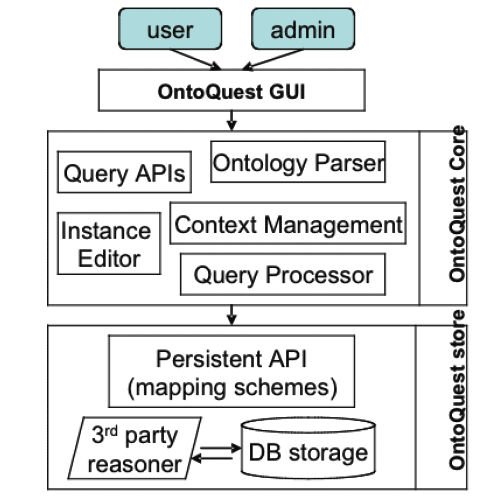
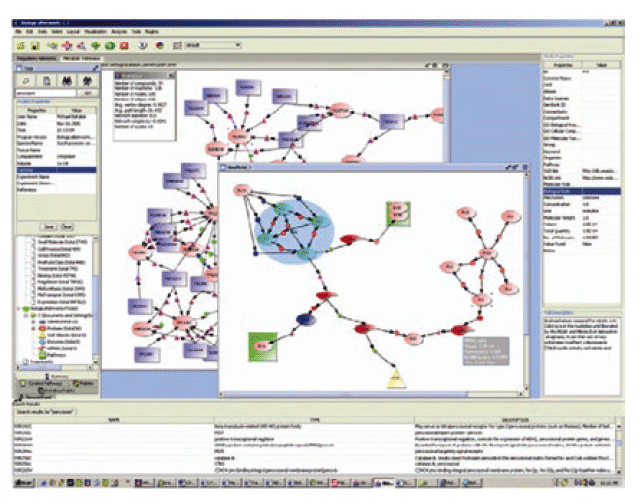
PathSys is a general-purpose, scalable, graph data warehouse of biological information, complete with a graph manipulation and a query language, a storage mechanism, and a generic data-importing mechanism through schema-mapping. The client software, named BiologicalNetworks, supports the navigation and analyses of molecular networks (Figure 9B). As systems biology integrates information from many data sources, distributed or integrated, there is an increasing demand for managing, querying and automated reasoning using ontology concepts and description logic. OntoQuest is developed to provide the extended mapping schemes for storing OWL (web ontology language) ontologies into backend databases (Figure 9A). It is aimed to guide a user to explore ontological datasets and eventually make non-preconceived, impromptu discoveries.
Both PathSys and OntoQuest take advantages a new semantic-aware RDF (resource description framework) algebra, which supports the inference of complex relationships represented in ontological hierarchies. OntoQuest is being developed using the Cell Centered Database (CCDB) and neuroscience ontology, whereas PathSys is driven by research in yeast signaling pathways. The technology is extensible to the data integration between distributed data and computation services described with appropriate semantics and ontology terms (Figure 9). The technology being developed under OntoQuest may also be used to provide additional semantic annotations to Opal based web services to improve automated service discovery and utilization.