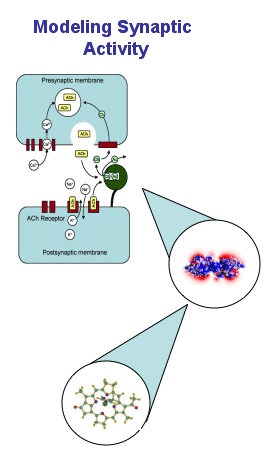
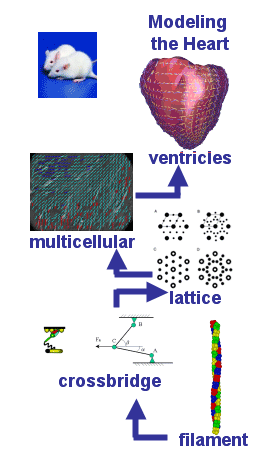
With the increasing availability of genome sequencing data, it is becoming apparent that knowing the parts is only a prerequisite to understand the big picture. While genomics and proteomics efforts are producing data at an increasing rate, the data are more of a descriptive nature rather than functional integration and interactions of the parts.1 Traditionally, most research and modeling activities have focused on a particular system level such as proteins, cells, tissues, organs, organ systems, up to the level of populations. Multiscale modeling, across the length scale from nanometers for molecules to meters for human bodies, as well as across the time scale from nano-seconds for molecular interactions to the length of human life, is crucial to the development of simulation systems for better understanding of human physiology and predictive capabilities for disease prevention and treatment.2
Multiscale modeling studies derive mathematical models of structure-function relations at one scale and link to the level below through appropriate parameters. These models need to be based on widely adopted modeling standards, with necessary software tools for developing, visualizing and linking the models.3 Multiscale modeling requires constant cross validation and feedback from experiments and models. Often the experiments provide the data for development and validation of the models, and the models can in turn provide predictions of behavior or require additional experiments which may lead to new discoveries.1 Models may impose a priori physical constraints and represent complex processes, and provide quantitative predictions that may be verified experimentally.4 Systems modeling are data-limited when mechanistic models are to be built, because experiments may be slow and difficult to validate. On the other hand, models across scales are compute-limited due to the “tyranny of scale”. Molecular dynamics simulations are often limited to a scale 5 to 6 orders of magnitude smaller than the time necessary for a real event to complete. In terms of an extreme case of computational challenge, the panel on Simulation Based Engineering Science (SBES)5 noted that in the turbulence-flow modeling, the “tyranny of scale” prevents a solution for many generations, even with Moore’s law holding true.
In recognition of the importance and the severe obstacles to experiments that are cross-scale in space, time and state, there have been a number of workshops held and panel recommendations made. The multiscale modeling consortium or Interagency Modeling and Analysis Group (IMAG),6 along with the participation of a number of federal agencies that the National Institutes of Health (NIH), National Science Foundation (NSF), National Aviation and Space Agency (NASA), Depart of Energy, (DOE), Department of Defense (DoD) and the United States Department of Agriculture (USDA), aims to promote the development and exchange of tools, models, data and standards for the MultiScale Modeling (MSM) community. The NSF blue ribbon panel 5 recognizes that SBES applied to the multiscale study of biological systems and clinical medicine, or simulation based medicine, may bring us closer to the realization of P4 medicine (predictive, preventative, personalized, and participatory). The June 2005 PITAC report on computational science7 and the 2005 National Research Council report,8 both specifically recommended increased and sustained support for infrastructure development to meet the computational challenges ahead.
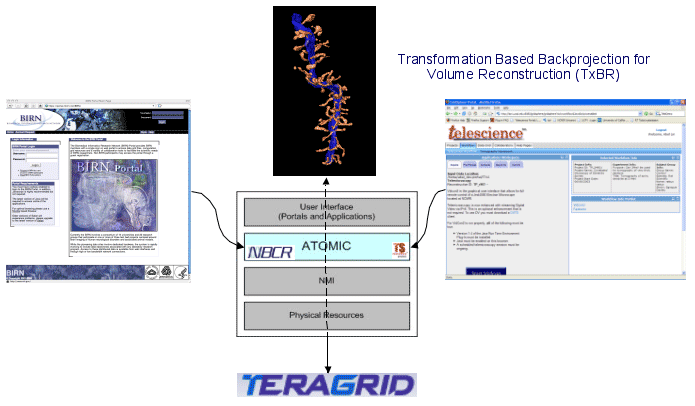
The National Biomedical Computation Resource (NBCR9), a national center supported by the National Center for Research Resources (NCRR10), has a mission to “conduct, catalyze and enable multiscale biomedical research” by harnessing advanced computation and data cyberinfrastructure through multidiscipline and multi-institutional integrative research and development activities (Figure 1). NBCR has co-hosted several workshops and conferences that engage researchers and students from the MSM community.11 12 13 It has been our experience that tools developed with one application in mind tend to be narrow and less flexible, let alone interoperable, with other software. Therefore, we have been focusing on enabling a set of pathfinder examples across scales, through team work with world class scientists who are members of NBCR, and through collaborative projects with members of the broader biomedical and MSM communities.