
August 2006
|
National Biomedical Computation Resource (NBCR): Developing End-to-End Cyberinfrastructure for Multiscale Modeling in Biomedical Research
|
 August 2006 |
Abstract — Begun in 1994, the mission of the National Biomedical Computation Resource (NBCR) is to conduct, catalyze and enable multiscale biomedical research by harnessing advanced computation and data cyberinfrastructure through multidiscipline and multi-institutional integrative research and development activities. Here we report the more recent research and technology advances in building cyberinfrastructure for multiscale modeling activities.
The development of the cyberinfrastructure is driven by multiscale modeling applications, which focus on scientific research ranging in biological scale from the subatomic, to molecular, cellular, tissue to organ level. Application examples include quantum mechanics modeling with GAMESS, calculation of protein electrostatic potentials with APBS and the finite element toolkit FEtk; protein-ligand docking studies with AutoDock; cardiac systems biology and physiology modeling with Continuity; and molecular visualizations using PMV and visual workflow programming in Vision. Real use cases are used to demonstrate how these multiscale applications may be made available transparently on the grid to researchers in the biomedicine and translational research arena, through integrative projects ranging from the understanding of the detailed mechanism of HIV protease and integrase action, to neuromuscular junction research in myopathy, to heart arrhythmia and failure, and to emerging public health threats, as well as through collaborative projects with other research teams across the world.
The adoption of service oriented architecture enables the development of highly reusable software components and efficiently leverages the international grid development activities. We describe an end to end prototype environment, exemplified by the adoption of key components of the Telescience project, that allows existing applications to run transparently on the grid, taking advantage of open source software that provides the following:
Solutions to complex problems may be developed using workflow tools that coordinate different interoperable services. In addition, we also describe the development of ontology and semantic mediation tools such as Pathsys and OntoQuest for data integration and interoperability, which may be efficiently coupled with the application services provided to the biomedical community.
With the increasing availability of genome sequencing data, it is becoming apparent that knowing the parts is only a prerequisite to understand the big picture. While genomics and proteomics efforts are producing data at an increasing rate, the data are more of a descriptive nature rather than functional integration and interactions of the parts.1 Traditionally, most research and modeling activities have focused on a particular system level such as proteins, cells, tissues, organs, organ systems, up to the level of populations. Multiscale modeling, across the length scale from nanometers for molecules to meters for human bodies, as well as across the time scale from nano-seconds for molecular interactions to the length of human life, is crucial to the development of simulation systems for better understanding of human physiology and predictive capabilities for disease prevention and treatment.2
Multiscale modeling studies derive mathematical models of structure-function relations at one scale and link to the level below through appropriate parameters. These models need to be based on widely adopted modeling standards, with necessary software tools for developing, visualizing and linking the models.3 Multiscale modeling requires constant cross validation and feedback from experiments and models. Often the experiments provide the data for development and validation of the models, and the models can in turn provide predictions of behavior or require additional experiments which may lead to new discoveries.1 Models may impose a priori physical constraints and represent complex processes, and provide quantitative predictions that may be verified experimentally.4 Systems modeling are data-limited when mechanistic models are to be built, because experiments may be slow and difficult to validate. On the other hand, models across scales are compute-limited due to the “tyranny of scale”. Molecular dynamics simulations are often limited to a scale 5 to 6 orders of magnitude smaller than the time necessary for a real event to complete. In terms of an extreme case of computational challenge, the panel on Simulation Based Engineering Science (SBES)5 noted that in the turbulence-flow modeling, the “tyranny of scale” prevents a solution for many generations, even with Moore’s law holding true.
In recognition of the importance and the severe obstacles to experiments that are cross-scale in space, time and state, there have been a number of workshops held and panel recommendations made. The multiscale modeling consortium or Interagency Modeling and Analysis Group (IMAG),6 along with the participation of a number of federal agencies that the National Institutes of Health (NIH), National Science Foundation (NSF), National Aviation and Space Agency (NASA), Depart of Energy, (DOE), Department of Defense (DoD) and the United States Department of Agriculture (USDA), aims to promote the development and exchange of tools, models, data and standards for the MultiScale Modeling (MSM) community. The NSF blue ribbon panel 5 recognizes that SBES applied to the multiscale study of biological systems and clinical medicine, or simulation based medicine, may bring us closer to the realization of P4 medicine (predictive, preventative, personalized, and participatory). The June 2005 PITAC report on computational science7 and the 2005 National Research Council report,8 both specifically recommended increased and sustained support for infrastructure development to meet the computational challenges ahead.
The National Biomedical Computation Resource (NBCR9), a national center supported by the National Center for Research Resources (NCRR10), has a mission to “conduct, catalyze and enable multiscale biomedical research” by harnessing advanced computation and data cyberinfrastructure through multidiscipline and multi-institutional integrative research and development activities (Figure 1). NBCR has co-hosted several workshops and conferences that engage researchers and students from the MSM community.11 12 13 It has been our experience that tools developed with one application in mind tend to be narrow and less flexible, let alone interoperable, with other software. Therefore, we have been focusing on enabling a set of pathfinder examples across scales, through team work with world class scientists who are members of NBCR, and through collaborative projects with members of the broader biomedical and MSM communities.
Understanding the workings of cells, tissues, organs, or entire organisms requires researchers to pull together information from multiple physical scales and across multiple temporal scales. We highlight activities within our collective and collaborative experiences at different or cross scales. These scientific examples are from long running complex multiscale research areas, which drive the development of our technology integration and infrastructure development efforts.
The need for integrative analysis in cardiac physiology and pathophysiology is readily appreciated. Common heart diseases are multi-factorial, multi-genic, and linked to other systemic disorders such as diabetes, hypertension, or thyroid disease. The coupling between the L-type Calcium channels (LCCs, also known as dihydropyridine receptors, or DHPRs) and ryanodine receptors (RyRs) are important in the excitation-contraction (E-C) of cardiac myocytes. The influx of calcium releases the calcium store in the Sarcoplasmic reticulum (SR), a phenomenon known as the calcium induced calcium release (CICR). In fact, the latest ionic models of cardiac myocytes include more than 20 ionic fluxes and 40 ordinary differential equations.1 Computational methods and ionic models for cardiac electromechanics at different scales have also been developed and are available in the software package Continuity. Figure 2 shows an example of how Continuity is used to help develop dual pacemaker systems that are helping to save people’s lives today 14. Continuity is used by a number of researchers in the field of cardiac biomechanics, and receives regular acknowledgment in peer-reviewed publications.15 16 Continuity 6 is continuously being improved to support larger scale simulations, for example using the MYMPI package,17 a standards-driven MPI libraries for Python developed by NBCR to improve the parallel computation efficiency.
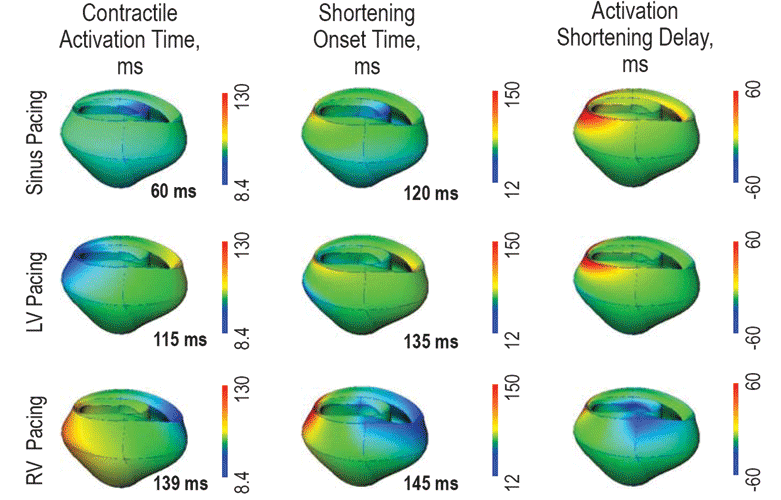
In ventricular myocytes, the dyadic cleft is a periplasmic space that spans about 10 nm between the voltage-gated LCCs/DHPRs on the transverse tubule (TT) membrane, and RyRs on the SR. Within the dyadic cleft, the small reaction volume and the exceedingly low number of reactant molecules means that the reaction system is better described by stochastic behavior, rather than continuous, deterministic reaction-diffusion “partial differential equations.”18 19 This system is one of the focal points of NBCR research at the molecular scale, and for cross scale integrations, with the development of highly realistic 3D models based on electron tomography from the National Center for Microcopy and Imaging Research (NCMIR).
The Python Molecular Viewer (PMV) is a component based software package (Figure 3) written in Python and contains an accompanying visual programming tool called Vision.20 PMV is among the first molecular visualization software packages that takes advantage of grid services, using the web service toolkit Opal developed by NBCR, to access remote databases and computational resources (Figure 3A). Many key packages from the PMV/Vision framework have been reused in Continuity 6, the multiscale modeling platform for cardiac electrophysiology and biomechanics.
In addition, AutoDock Tools (ADT) 21 has been developed as a module inside PMV for the popular molecular docking software package AutoDock. AutoDock is a world famous software package and has been used in developing inhibitors for many important diseases.22 23 The FightAids@Home project has been using AutoDock to screen for HIV inhibitors and is now running on the World Community Grid, an IBM philanthropic activity.24 ADT greatly simplifies the preparation and post analysis procedures of AutoDock (Figure 3B).
The use of computer models to screen for small molecules that will bind to and alter the activity of macromolecules has long been complicated by “induced fit” effects: the macromolecule may undergo shape changes during the formation of the complex with the small molecule. A new approach has been developed in which a variety of conformations of the target macromolecule are generated by molecular dynamics simulation or other methods, to generate structures that might include one or more that are representative of that of the “relaxed complex” with the small molecule.25 To select the most relevant macromolecular conformations, rapid screening is done using AutoDock and ADT. The most stable complex structures from this screening are then subject to more refined analysis to yield the most probable structure of the complex and an associated estimate of the strength of binding (Figure 3C). More recently, applications of this method to an antiviral target for treatment of HIV infections 26 has aided scientists at Merck & Co. in developing a new class of drugs that are in Phase III clinical trials.27
The Finite Element Tool Kit (FEtk)28 is an evolving collection of parallel adaptive, multilevel finite element software libraries and supports tools for solving coupled systems of partial differential equations (PDE) and integral equations (IE).29 The numerical libraries are written in an object-oriented form of ANSI-C (Figure 4A). Left (Top/Bottom): Potential contours of the electrostatic potential around a biomolecule, computed adaptively using FEtk, and a closeup of the adapted part of the simplex mesh. Right (Top/Bottom): Isosurfaces projected onto a cutting plane through two black holes in an astrophysics problem. FEtk was used to compute the initial bending of space and time around two massive black holes, which involved adaptively solving a coupled nonlinear elliptic PDE.
The primary FEtk ANSI-C software libraries include MALOC (Minimal Abstraction Layer for Object-oriented C) for portability, SG (Socket Graphics) for networked OpenGL-based visualization, and MC (Manifold Code) for adaptively solving coupled systems of nonlinear PDE on two- and three-manifold domains. A 2D MATLAB-based prototyping tool called MCLite is also available for fast development of MC-based software. A related package, PMG (Parallel Algebraic Multigrid) is designed to numerically solve scalar nonlinear PDE problems such as the Poisson-Boltzmann Equation (PBE) on a class of regular domains using algorithms which have optimal or near-optimal space and time complexity.
APBS is a software package for the numerical solution of the PBE, one of the most popular continuum models for describing electrostatic interactions between molecular solutes in salty, aqueous media.30 Continuum electrostatics plays an important role in several areas of biomolecular simulation, including:
APBS was designed to efficiently evaluate electrostatic properties for such simulations for a wide range of length scales to enable the investigation of molecules with tens to millions of atoms. APBS uses FEtk and PMG to solve the Poisson-Boltzmann equation numerically.
APBS is available to users as standalone applications or inside PMV for seamless access (Figure 4B), available in CHARMM or AMBER through the NBCR iAPBS interface,31 or as web services using the NBCR Opal toolkit.32 In this example, the electrostatic binding energy for the protein kinase A (PKA) complex with the inhibitor balanol was computed using APBS and displayed in PMV. The PKA complex is shown as a ribbon diagram with side chains or positively (blue) and negatively (red) charged amino acids displayed as Sticks and Balls. The inhibitor molecule is displayed using a fat Sticks and Balls representation and colored by atom types. The electrostatic binding energy is visualized by direct volume rendering with the shown transfer function and using two isosurfaces.
A new software component from NBCR that allows the efficient modeling of diffusion events across the neuromuscular junction in a steady state or time dependent manner called the Smoluchowski equation solver (SMOL) is also based on the FEtk and has been used in conjunction with APBS in a study of the tetrameric complex acetylcholine receptors.33
In parallel to efforts focusing attention on the needs and the benefits of multiscale modeling, tremendous national and international investments have already been made to develop and deploy a cyberinfrastructure that will revolutionize the conduct of science.34 This cyberinfrastructure consists of distributed computational, data storage, observational, and visualization resources, including human resources, connected by a network infrastructure and a software layer (middleware), that will “bring access of resources (at one end) to researchers (at another) and allow researchers to conduct team science as part of normal conduct of science 35, in an end-to-end cyberinfrastructure.” Cyberinfrastructure, and grid, are often used interchangeably.36
The necessary grid infrastructure to support the multiscale modeling community remains to be defined through an iterative process with ongoing interactions between scientists and infrastructure developers. There has been significant progress in the development of the networks and physical resources that are the fabric of the grid.37 However, the middleware layer, which connects the fabric with the users and applications, is still in a state of flux. To increase the usability and decrease the cost of entry of the grid, new programming models or application execution environments are being developed, and sometimes these are referred to as grid application-level tools.38 These tools are designed to be built on top of the grid software infrastructure. They are generic, easy to use and shield the users from changes in underlying architecture.
An “end-to-end” cyberinfrastructure for multiscale modeling needs to have the capability to handle the representative data commonly encountered in MSM, by representative users who need to accomplish tasks that are representative of the nature of MSM research and development. The scientific drivers for multiscale modeling at NBCR are diverse and span the scale from subatomic electron charge density flows in development of better photodynamic therapies 39, to cellular and whole heart physiology modeling,1 with information integration technology providing the mediator necessary for interoperability. All these projects drive the development and integration effort we are leading to provide the cyberinfrastructure necessary and required for the research objectives to be met. In addition, NBCR works closely with other projects at UCSD, such as NCMIR, BIRN,40 CAMERA,41 Optiputer,42 JCSG,43 and establishes important collaborations with national projects such as TeraGrid,44 Open Science Grid,45 as well as international projects such as PRAGMA46 and OMII.47
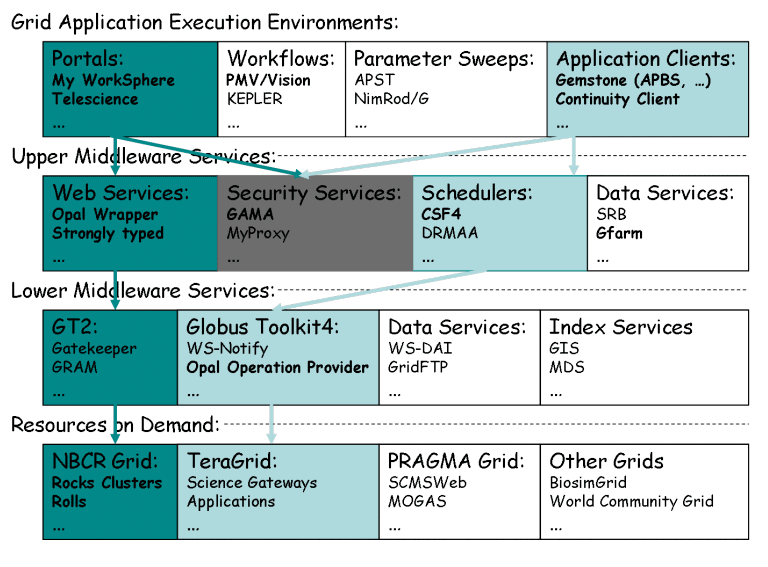
The development of infrastructure that can support diverse applications using distributed physical resources and remain easy to use and scalable has ushered in the service-oriented architecture as the dominant modus operandi for reasons eloquently stated in.48 To enable scientific applications on the grid, many different approaches have been adopted.49 We have taken a minimalist approach, which is to select the most stable components, achieve the greatest leverage, and develop smart glues that are reusable components, within the service oriented approach (Figure 5). We’ll highlight some key components developed by or with critical contributions from NBCR and then discuss how they are used to support multiscale modeling efforts at NBCR and how they are available to the MSM community in general.
Upper middleware services are those that make the development of distributed applications significantly easier, with support for higher levels of abstraction and standardization. For example, NBCR Opal based application web services provide job management and scheduling features based on the Globus toolkit. An application developer may begin using the grid quickly with the basic knowledge of web service development, as shown by the use cases in PMV, My WorkSphere, and Gemstone user environments. Lower middleware services are those that have stabilized over the years and serve as the foundation for the development of more sophisticated and transparent modes of access. However, as often dictated by performance requirements, a user application may access lower layers directly. This is much less desirable unless the integration is based on the service oriented architecture. Below we describe some key tools and technology actively developed or co-developed by NBCR to support our user communities. For a more comprehensive description of all the tools and software, please see NBCR download site.50
Opal is developed by NBCR as a Java based toolkit that automatically wraps any legacy applications with a Web services layer that is fully integrated with Grid Security Infrastructure (GSI) based security, cluster support, and data management.32 51 The advantage of using OPAL is that the application may be launched using any Web service client, because the WSDL defines using the standard protocol for how the service may be accessed, and it provides the basic HTTP access to results, as well as any metadata that describes how the results may be handled (Figure 6A). This ‘deploy once use by many’ feature of a Web service is a key ingredient for achieving interoperability. Because OPAL manages the interaction with the grid architecture, the grid is transparent to the user. In addition, workflow tools like Kepler52 may easily compose web services based workflows through a common interface (Figure 6B).
GAMA, co-developed by NBCR,53 is a GSI-based security service that manages X.509 user credentials on behalf of users and supports SOAP-based applications (Figure 7A). The server component leverages existing software packages such as CAS, MyProxy, and CACL with the GAMA version 2 supporting other CA packages such as NAREGI. A portlet component provides the administrative interface to the server. As security is a sensitive and critical issue in the production use of the cyberinfrastructure, GAMA allows any (Figure 7) organization to create their own certificate authority, manage their user certificates, secure the SOAP communications using HTTPS and mutual authentication, and integrate seamlessly with portals and rich clients. For example, GAMA is used by GridSphere based portals,54 Gemstone55 and PMV/Vision.
The ATOMIC bundle (Figure 7B), developed for the Telescience Project by NBCR, provides a fabric for seamless interoperability among user interfaces (web portals and applications) and externally addressable grid resources (instruments and computers) inside a grid portal environment.56 There is a unified programming API for basic scientific research, which liberates the users and programmers from having to learn about the grid. TeleAuth is a specialized version of GAMA. TeleWrap provides seamless access to remote data transparently and has been used successfully in the Telescience portal and EMWorkspace. TeleRun provides a higher level of abstraction than Opal-provided services and may further shield developers from the details of grid execution environment. ATOMIC also provides persistent session information during a particular Telescience session, so that all user portlets have access to the same session information regardless of the specific portlet being used.
The development of MYMPI57 is an effort of NBCR, driven by the requirement of Continuity, to improve interprocess communications between Python and FORTRAN.17 It is a Python module used with a normal Python interpreter. MyMPI is an implementation following the standard specifications of MPI, supporting more than 30 of the commonly used MPI calls. The syntax of the calls matches the syntax of C and FORTRAN calls closely to support mixed Python with the other languages. MYMPI will also allow access from Python to other application packages such as the Finite Element Toolkit (FEtk).58
Gemstone59 is developed with major support from the NSF National Middleware Initiative, in close collaboration with NBCR, especially in the interaction with strongly typed and Opal based web services (Figure 8 A). For example, Gemstone provides an interface to GAMESS60 and APBS strongly typed web services,30 both developed by NBCR. It also supports the GARNET visualization web service from the NMI (NSF Middleware Initiative) project. Opal based web service for PDB2PQR,61 a utility package for APBS calculations, is also accessible from Gemstone as web service based workflows. Gemstone utilizes the open source Mozilla engine and uses XML User Interface Markup Language (XUL) to describe the user interface. My WorkSphere is a prototype GridSphere based portal environment, which leverages portlets that are JSR168 standard compliant. As an example, we have deployed MEME62 as a portlet using Opal, as part of a generalized Cyberinfrastructure for bioinformatics applications,51 with Gfarm63 and CSF464 as the other key middleware components.
In an effort to make cyberinfrastructure more readily available to scientists and engineers, it is necessary to not only develop different middleware to support legacy applications, but also to make the different software packages easy to deploy into existing infrastructure. Rocks cluster environment toolkit65 (Figure 8B) has proven to be invaluable for NBCR to build the basic infrastructure, deploy our software stack, and make our infrastructure replicable by others. NBCR has contributed critically to the development of the Condor roll66 (a roll is a mechanism, similar to the Red Hat RPM, though fully automated, for building reproducible cluster and grid environments). Other rolls available from NBCR include the APBS, MEME, GAMA, AutoDock, and PMV. Additional rolls for SMOL67 and FEtk will be available soon (Figure 8 B).
PathSys is a general-purpose, scalable, graph data warehouse of biological information, complete with a graph manipulation and a query language, a storage mechanism, and a generic data-importing mechanism through schema-mapping. The client software, named BiologicalNetworks, supports the navigation and analyses of molecular networks (Figure 9B). As systems biology integrates information from many data sources, distributed or integrated, there is an increasing demand for managing, querying and automated reasoning using ontology concepts and description logic. OntoQuest is developed to provide the extended mapping schemes for storing OWL (web ontology language) ontologies into backend databases (Figure 9A). It is aimed to guide a user to explore ontological datasets and eventually make non-preconceived, impromptu discoveries.
Both PathSys and OntoQuest take advantages a new semantic-aware RDF (resource description framework) algebra, which supports the inference of complex relationships represented in ontological hierarchies. OntoQuest is being developed using the Cell Centered Database (CCDB) and neuroscience ontology, whereas PathSys is driven by research in yeast signaling pathways. The technology is extensible to the data integration between distributed data and computation services described with appropriate semantics and ontology terms (Figure 9). The technology being developed under OntoQuest may also be used to provide additional semantic annotations to Opal based web services to improve automated service discovery and utilization.
There are a plethora of tools and innovative approaches to the development of cyberinfrastructure in order to support multiscale modeling activities. However, better and more robust approaches will always come out of close collaborations between computer scientists and biomedical researchers, as well as other field specialists. The interactions will educate all the groups to be fully aware of the requirements and challenges of the state of the art technology, and make routine use of the grid possible today. In addition, the development of new tools that support applications in different fields and through international collaborations greatly reduces the collective cost for global computational grids. The service oriented approach is gaining momentum and greatly facilitates the development of a knowledge based global economy.
There are often conflicting requirements between biomedical research and grid communities, with the former accustomed to a “one experiment at a time” approach, and the latter desiring systems that handle large number of jobs simultaneously. The reality is that current grid computing systems are still difficult to learn, only relatively stable, and limited by the technologies available in hardware, software, and programming languages, as noted in [3]. It is a challenge to both communities to design better software and use them effectively. With these caveats in mind, one may attempt to select from the available tools and build a robust platform to make routine use of the grid possible, through close collaborations with developers of all components involved where possible. By working with different applications and addressing the common needs and individual requirements, reusable components may be isolated without sacrificing the customized environments demanded by users. The structure of resources supported by the National Center for Research Resources, such as NBCR and NCMIR, requires collaborative projects to guide the development of the tools, as noted above.
The number of tools showed in Figure 5, along with those represented only as “…”, offer many possible combinations to build end to end problem solving environments, often with overlapping features. For every tool listed, there are solid alternatives under different use cases. As shown in the usage scenarios, the ultimate choice of tools depends on the specific problems to be solved and the target audience of the designed environment. As demonstrated by the number of tools that are using Opal based services, the service oriented approach provides the flexibility in customized front-end tools with transparent access to underlying distributed computation resources. The demands of multiscale modeling applications will drive the development of the computation and data cyberinfrastructure, which in turn enables simulation based biology and medicine. The ensuing challenges in the search for solutions of more difficult problems will further this iterative developmental cycle of technology and science, with continuous and immediate impact to the health and well being of the public.
In developing collaborations and ultimately a user base to conduct research, having a persistent infrastructure is required. By this we mean access to tools and software for researchers to experiment with. The National Centers for Research Resources provide that type of infrastructure for the community. In the case of NBCR, access to the tools and services discussed in this article can be found at the NBCR 9 website.
While the experiences and developments discussed above have been informed by our local activities, the concepts of cyberinfrastructure are not bound by national borders. Examples of large scale projects and smaller team projects have been commented on in prior CTWatch issues (e.g., February 2006). Furthermore, it is essential that we actively participate in the international arena, both to ensure the various CI efforts can interoperate (e.g., when we want to use unique resources in other countries) and to reduce reproduction of tools that already exist.
Another exciting development that will link biomedical research with the environment across various scales is the newly funded CAMERA project. CAMERA will provide a community resource that links genetic information from a metagenomics effort, the Global Ocean Survey, with environmental factors such location, temperature and chemistry of the sampled environment.
The critical role played by NBCR to bridge the multiscale modeling research community and cyberinfrastructure is an important one, and will require collaborative efforts from other projects to expedite the translational/medical potential of multiscale biomedical and cyberinfrastructure research.