Understanding the workings of cells, tissues, organs, or entire organisms requires researchers to pull together information from multiple physical scales and across multiple temporal scales. We highlight activities within our collective and collaborative experiences at different or cross scales. These scientific examples are from long running complex multiscale research areas, which drive the development of our technology integration and infrastructure development efforts.
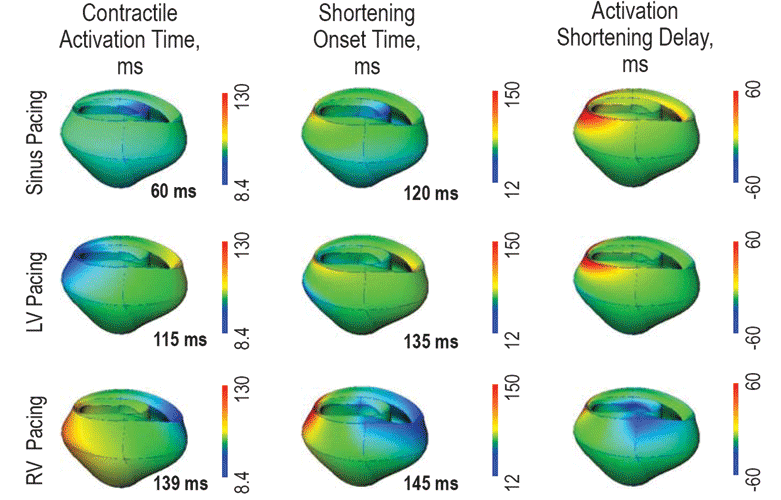
The need for integrative analysis in cardiac physiology and pathophysiology is readily appreciated. Common heart diseases are multi-factorial, multi-genic, and linked to other systemic disorders such as diabetes, hypertension, or thyroid disease. The coupling between the L-type Calcium channels (LCCs, also known as dihydropyridine receptors, or DHPRs) and ryanodine receptors (RyRs) are important in the excitation-contraction (E-C) of cardiac myocytes. The influx of calcium releases the calcium store in the Sarcoplasmic reticulum (SR), a phenomenon known as the calcium induced calcium release (CICR). In fact, the latest ionic models of cardiac myocytes include more than 20 ionic fluxes and 40 ordinary differential equations.1 Computational methods and ionic models for cardiac electromechanics at different scales have also been developed and are available in the software package Continuity. Figure 2 shows an example of how Continuity is used to help develop dual pacemaker systems that are helping to save people’s lives today 14. Continuity is used by a number of researchers in the field of cardiac biomechanics, and receives regular acknowledgment in peer-reviewed publications.15 16 Continuity 6 is continuously being improved to support larger scale simulations, for example using the MYMPI package,17 a standards-driven MPI libraries for Python developed by NBCR to improve the parallel computation efficiency.
In ventricular myocytes, the dyadic cleft is a periplasmic space that spans about 10 nm between the voltage-gated LCCs/DHPRs on the transverse tubule (TT) membrane, and RyRs on the SR. Within the dyadic cleft, the small reaction volume and the exceedingly low number of reactant molecules means that the reaction system is better described by stochastic behavior, rather than continuous, deterministic reaction-diffusion “partial differential equations.”18 19 This system is one of the focal points of NBCR research at the molecular scale, and for cross scale integrations, with the development of highly realistic 3D models based on electron tomography from the National Center for Microcopy and Imaging Research (NCMIR).
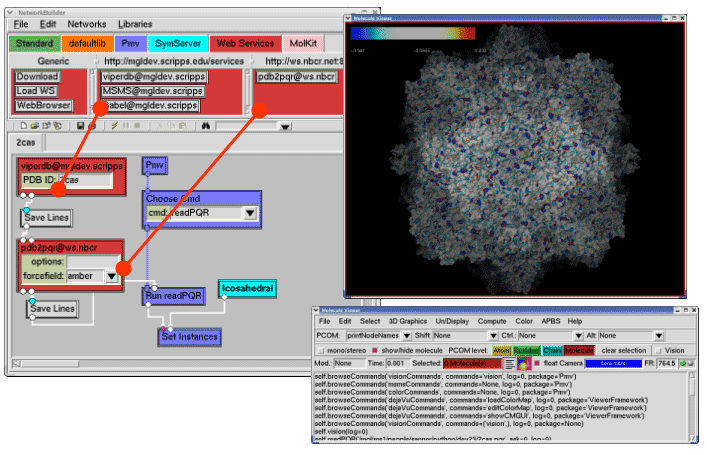
The Python Molecular Viewer (PMV) is a component based software package (Figure 3) written in Python and contains an accompanying visual programming tool called Vision.20 PMV is among the first molecular visualization software packages that takes advantage of grid services, using the web service toolkit Opal developed by NBCR, to access remote databases and computational resources (Figure 3A). Many key packages from the PMV/Vision framework have been reused in Continuity 6, the multiscale modeling platform for cardiac electrophysiology and biomechanics.
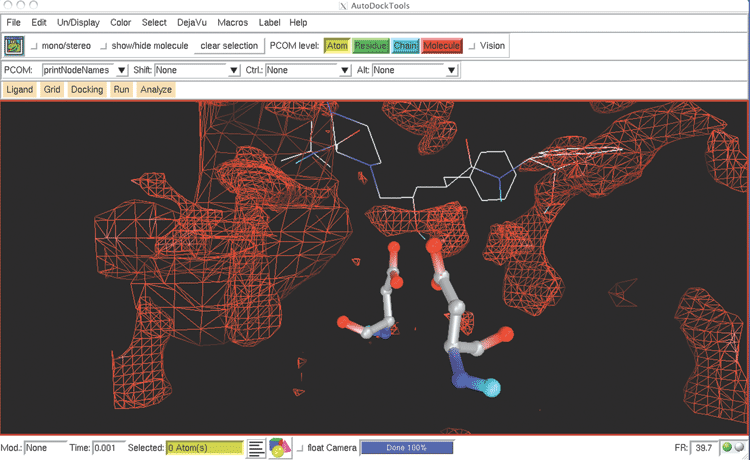
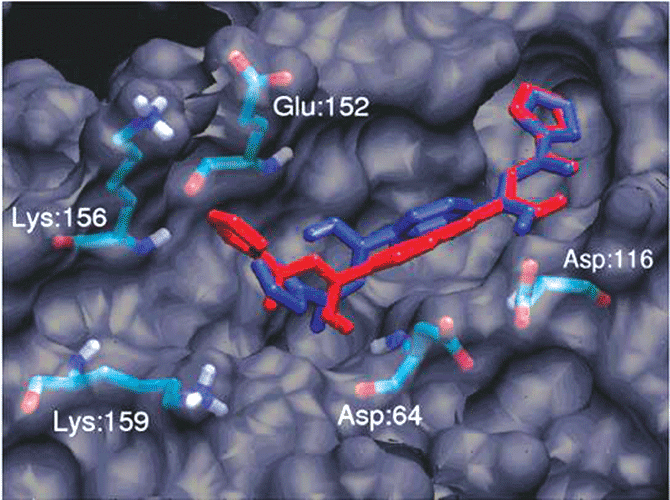
In addition, AutoDock Tools (ADT) 21 has been developed as a module inside PMV for the popular molecular docking software package AutoDock. AutoDock is a world famous software package and has been used in developing inhibitors for many important diseases.22 23 The FightAids@Home project has been using AutoDock to screen for HIV inhibitors and is now running on the World Community Grid, an IBM philanthropic activity.24 ADT greatly simplifies the preparation and post analysis procedures of AutoDock (Figure 3B).