Volume 2 Number 3
August 2006Trends and Tools in Bioinformatics and Computational Biology
Wilfred W. Li, University of California, San Diego (UCSD), San Diego Supercomputer Center (SDSC)
Nathan Baker, Washington University in Saint Louis
Kim Baldridge, UCSD, SDSC
J. Andrew McCammon, UCSD
Mark H. Ellisman, UCSD, Center for Research In Biological Systems (CRBS)
Amarnath Gupta, UCSD, SDSC
Michael Holst, UCSD
Andrew D. McCulloch, UCSD
Anushka Michailova, UCSD
Phil Papadopoulos, UCSD, SDSC
Art Olson, The Scripps Research Institute (TSRI)
Michel Sanner, TSRI
Peter W. Arzberger, California Institute for Telecommunications and Information Technology (Calit2), CRBS, UCSD
4
2.3. Relaxed Complex Method and its Application in HIV Integrase Inhibitor Studies
The use of computer models to screen for small molecules that will bind to and alter the activity of macromolecules has long been complicated by “induced fit” effects: the macromolecule may undergo shape changes during the formation of the complex with the small molecule. A new approach has been developed in which a variety of conformations of the target macromolecule are generated by molecular dynamics simulation or other methods, to generate structures that might include one or more that are representative of that of the “relaxed complex” with the small molecule.25 To select the most relevant macromolecular conformations, rapid screening is done using AutoDock and ADT. The most stable complex structures from this screening are then subject to more refined analysis to yield the most probable structure of the complex and an associated estimate of the strength of binding (Figure 3C). More recently, applications of this method to an antiviral target for treatment of HIV infections 26 has aided scientists at Merck & Co. in developing a new class of drugs that are in Phase III clinical trials.27
| A |
|
| B |
|
|
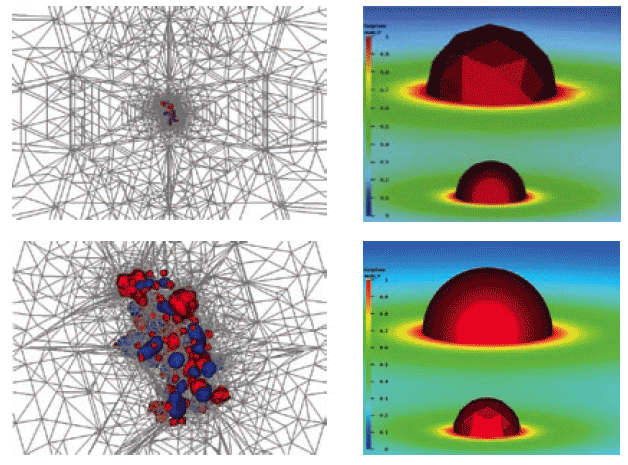
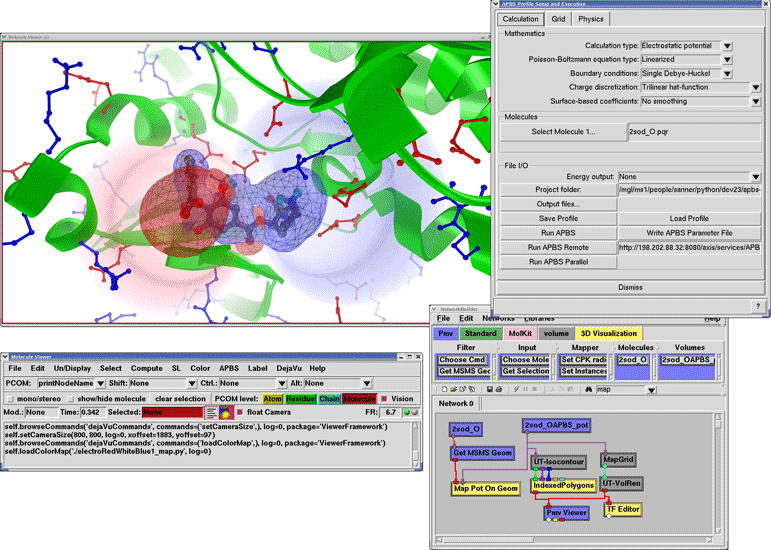
Figure 4. A) Application examples of FEtk for solving the electrostatic potentials of biomolecules or rendering of isosurfaces of black holes in astrophysics problems. B) The electrostatic binding energy of PKA and balanol is visualized using PMV after remote distributed APBS calculations using NBCR strongly typed web service.
|
Pages: 1 2 3 4 5 6 7 8 9 10 11
Reference this articleLi, W. W., Baker, N., Baldridge, K., McCammon, J. A., Ellisman, M. H., Gupta, A., Holst, M., McCulloch, A. D., Michailova, A., Papadopoulos, P., Olson, A., Sanner, M., Arzberger P. W. "National Biomedical Computation Resource (NBCR): Developing End-to-End Cyberinfrastructure for Multiscale Modeling in Biomedical Research,"
CTWatch Quarterly, Volume 2, Number 3,
August 2006. http://www.ctwatch.org/quarterly/articles/2006/08/national-biomedical-computation-resource/








