HemeLB is intended to yield patient-specific information, which helps plan embolisation of arterio-venous malformations and aneurysms, amongst other neuro-pathologies. Using this methodology, patient-specific models can be used to address issues with pulsatile blood flow, phase differences and the effects of treatment, all of which are potentially very powerful both in terms of understanding neurovascular patho-physiology and in planning patient treatment.
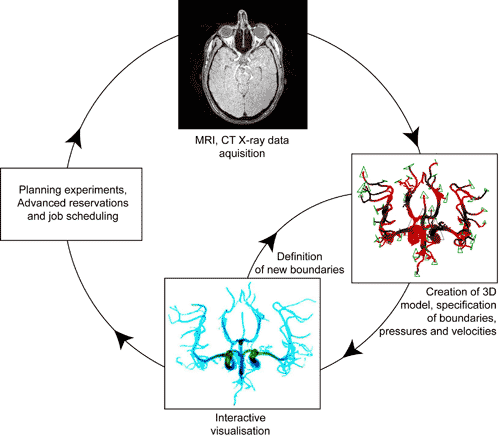
The software environment used in this project aims to bring to the forefront details and processes clinicians need to be aware of, such as (i) the process of image segmentation to obtain a 3D neurovascular model, (ii) the specification of pressure and velocity boundary conditions, and (iii) the real-time rendered image (Figure 2). Clinicians are not concerned with where simulations are running, nor the details of reservations, thus features such as advanced reservations and emergency computing capabilities, job launching and research selection are all done behind the scenes. This environment is particularly important given the time scales involved in the clinical decision making process in the treatment of aterio-venous malformations and aneurysms. From the acquisition of a 3D dataset (which is typically 2 to 4 GB in size), to the next embolisation, a time scale of 15 to 20 minutes is typical, and for such computational approaches to be clinically relevant, we have to fit into this time scale. There are also preventative scenarios that can be envisioned; patients could be subjected to such simulations in advance of vascular pathologies developing, averting future problems with interventional treatments.