
November 2007
|
Scientific Data Management: Essential Technology for Accelerating Scientific Discoveries
|
 November 2007 |
Terascale computing and large scientific experiments produce enormous quantities of data that require effective and efficient management. The task of managing scientific data is so overwhelming that scientists spend much of their time managing the data by developing special purpose solutions, rather than using their time effectively for scientific investigation and discovery. Effectively generating, managing, and analyzing this information requires a comprehensive, end-to-end approach to data management that encompasses all of the stages, from the initial data acquisition to the final analysis of the data. Fortunately, the data management problems encountered by most scientific domains are common enough to be addressed through shared technology solutions. Based on community input, we have identified three significant requirements. First, more efficient access to storage systems is needed. In particular, parallel file system improvements are needed to read and write large volumes of data without slowing a simulation, analysis, or visualization engine. These processes are complicated by the fact that scientific data are structured differently for specific application domains, and are stored in specialized file formats. Second, scientists require technologies to facilitate better understanding of their data, in particular the ability to effectively perform complex data analysis and searches over large data sets. Specialized feature discovery and statistical analysis techniques are needed before the data can be understood or visualized. To facilitate efficient access, it is necessary to keep track of the location of the datasets, effectively manage storage resources, and efficiently select subsets of the data. Finally, generating the data, collecting and storing the results, data post-processing, and analysis of results is a tedious, fragmented process. Tools for automation of this process in a robust, tractable, and recoverable fashion are required to enhance scientific exploration.
The Scientific Data Management (SDM) Center,1 funded under the DOE SciDAC program, focuses on the application of known and emerging data management technologies to scientific applications. The Center’s goals are to integrate and deploy software-based solutions to the efficient and effective management of large volumes of data generated by scientific applications. Our purpose is not only to achieve efficient storage and access to the data using specialized indexing, compression, and parallel storage and access technology, but also to enhance the effective use of the scientist’s time by eliminating unproductive simulations, by providing specialized data-mining techniques, by streamlining time-consuming tasks, and by automating the scientist’s workflows. Our approach is to provide an integrated scientific data management framework where components can be chosen by the scientists and applied to their specific domains. By overcoming the data management bottlenecks and unnecessary information-technology overhead through the use of this integrated framework, scientists are freed to concentrate on their science and achieve new scientific insights.
As part of our evolutionary technology development and deployment process (from research through prototypes to deployment and infrastructure) we have organized our activities in three layers that abstract the end-to-end data flow described above. We labeled the layers as Storage Efficient Access (SEA), Data Mining and Analytics (DMA), and Scientific Process Automation (SPA). The SEA layer is immediately on top of hardware and operating systems, providing parallel data access to files and transparent access to archival storage. The DMA layer, which builds on the functionality of the SEA layer, consists of indexing, feature selection, and parallel statistical analysis technology. The SPA layer, which is on top of the DMA layer, provides the ability to compose workflows from the components in the DMA layer as well as application specific modules. Figure 1 shows this organization and the components developed by the center and applied to various scientific applications.

Over the last several years, the technologies supported by the SDM center have been deployed for a variety of application domains. Some of the most notable achievements are:
In this section we describe the SDM Center technologies, and include some examples of their application in various scientific projects. We proceed with technologies from the top layer to the bottom layer.
A practical bottleneck for more effective use of available computational and data resources is often the design of resource access and use of processes, and the corresponding execution environments, i.e., in the scientific workflow environment of end user scientists. The goal of the Kepler system2 is to provide solutions and products for effective and efficient modeling, design and execution of scientific workflows. Kepler is a multi-site open source effort, co-founded by the SDM center, to extend the Ptolemy system (from UC Berkeley) and create an integrated scientific workflow infrastructure. We have also started to incorporate data, process, system and workflow provenance and run-time tracking and monitoring. We have worked closely with application scientists to design, implement, and deploy workflows that address their real-world needs. In particular, we have active users on the SciDAC Terascale Supernova Initiative (TSI) team and an LLNL Biotechnology project, as well as at the Center for Plasma Edge Simulation (CPES) fusion project. While the Scientific Process Automation (SPA) layer uses Kepler to achieve workflow automation, it is the specific task components (called “actors” in Kepler) developed by the SDM center that makes our work unique in it usefulness to scientific applications.

Underlying challenges related to simulations, data analysis and data manipulation include scalable parallel numerical algorithms for the solution of large, often sparse linear systems, flow equations, and large Eigen-value problems, running of simulations on supercomputers, movement of large amounts of data over large distances, collaborative visualization and computational steering, and collection of appropriate process and simulation related status and provenance information. This requires interdisciplinary teams of application scientists and computer scientists working together to define the workflows and putting them into the Kepler workflow framework. The general underlying “templates” are often similar across disciplines: large-scale parallel computations and steering (hundreds of processors, gigabytes of memory, hours to weeks of CPU time), data-movement and reduction (terabytes of data), visualization and analytics (interactive, retrospective, and auditable). An abstraction of this and its Kepler translation are illustrated in Figure 2 and 3 for a particular astrophysics project, call the Terascale Supernova Initiative (TSI).3 Figure 3 shows the capability of the Kepler system to represent hierarchically structured workflows. In the center of the figure there are four simple high-level tasks; each is expanded into lower level tasks that manage the detailed processes.

As part of the Data Mining and Analysis (DMA) layer, the SDM center is developing scalable algorithms for the interactive exploration of large, complex, multi-dimensional scientific data. By applying and extending ideas from data mining, image and video processing, statistics, and pattern recognition, we are developing a new generation of computational tools and techniques that are being used to improve the way in which scientists extract useful information from data.4 These tools were applied to problems in a variety of application areas, including separation of signals in climate data from simulations, the identification of key features in sensor data from the D-III-D Tokamak, and the classification and characterization of orbits in Poincaré plots in Fusion data.

A specific example of the effectiveness of such techniques is the identification of the movement of “blobs” in images from fusion experiments, using data from the National Spherical Torus Experiment (NSTX),5 shown in Figure 4. A blob is a coherent structure in the image that carries heat and energy from the center of the torus to the wall. Figure 5 shows bright blobs extracted from experimental images from the NSTX. The blobs are high energy regions. If they hit the torus wall that confines the plasma, it can vaporize. The figure shows movement of the blobs over time. A key challenge to the analysis is the lack of a precise definition for these structures. Figure 5 shows three consecutive images from an NSTX sequence. The original images are somewhat noisy and must first be processed to remove the noise. We have applied our background subtraction software to remove the quiescent background intensity in the sequences. Next, ambient background intensity, which is approximated by the median of the sequence, is removed, thus highlighting the blob regions, as shown in the second row of the figure. We then use image processing techniques to identify and track the blobs over time, as shown in the third row. The goal is to validate and refine the theory of plasma turbulence.

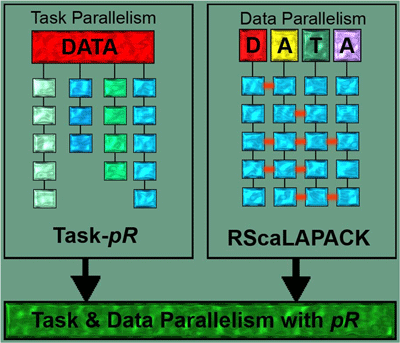
Another area supported by the DMA layer is efficient statistical analysis. Present data analysis tools such as Matlab, IDL, and R, even though highly advanced in providing various statistical analysis capabilities, are not apt to handle large data-sets. Most of the researchers’ time is spent on addressing data preparation and management needs of their analyses. Parallel R6 is an open source parallel statistical analysis package developed by the SDM center that lets scientists employ a wide range of statistical analysis routines on high performance shared and distributed memory architectures without having to deal with the intricacies of parallelizing these routines. Through Parallel R, the user can distribute data and carry out the required parallel computation but maintain the same look-and-feel interface of the R system. Two major levels of parallelism are supported: data parallelism (k-means clustering, Principal Component Analysis, Hierarchical Clustering, Distance matrix, Histogram) and task parallelism (Likelihood Maximization, Bootstrap and Jackknife Re-sampling, Markov Chain Monte Carlo, Animations). Figure 6 shows a schematic of the concepts. ParallelR has been applied in multiple scientific projects including feature extraction for quantitative high-throughput proteomics, parallel analyses of climate data, and in combination with geographical information systems.
Another aspect of effective data analysis supported by the DMA technology in the SDM center, is the ability to identify, in real-time, items of interest from billions of data values in large datasets. This is a significant challenge posed by the huge amount of data being produced by many data-intensive science applications. For example, a high-energy physics experiment called STAR is producing hundreds of terabytes of data a year and has accumulated many millions of files in the last five years of operation. One of the core missions of the STAR experiment is to verify the existence of a new state of matter called the Quark Gluon Plasma (QGP). An effective strategy for this task is to find the high-energy collisions that contain signatures unique to QGP, such as a phenomenon called jet quenching. Among the hundreds of millions of collision events captured, a very small fraction of them (maybe only a few hundreds) contain clear signatures of jet quenching. Efficiently identifying these events and transferring the relevant data files to analysis programs are a great challenge. Many data-intensive science applications are facing similar challenges in searching their data.
Over the last several years, we have been working on a set of strategies to address this type of searching problem. Usually, the data to be searched are read-only. Our approach takes advantage of this fact. We have developed a specialized indexing method based on representing the indexed data as a compressed bitmap. This indexing method, called FastBit,7 is an extremely efficient bitmap indexing technology. Unlike other bitmap indexes that assume low cardinality of possible data values, FastBit is particularly useful for scientific data, since it is designed for high-cardinality numeric data. FastBit performs 12 times faster than any known compressed bitmap index in answering range queries. Because of its speed, Fastbit facilitates real-time analysis of data, searching over billions of data values in seconds. FastBit has been applied to several application domains, including finding flame fronts in combustion data, searching for rare events from billions of high energy physics collision events, and more recently to facilitate query-based visualization. The examples in Figure 7 (for astrophysics and combustion data) show the use of a tool, called DEX,8 that used Fastbit in combination with VTK to achieve a very fast selection of features from large datsets and their display in real-time.


As high-performance computing applications scale and move from performing simulation and computing to data analysis they become tremendously data-intensive, creating a potential bottleneck in the entire scientific discovery cycle. At the same time, it is a well-known phenomenon that I/O access rates have not kept pace with high-performance computing performance as a whole. Because of this phenomenon, it becomes increasingly important for us to extract the highest possible performance from the I/O hardware that is available to us. Even if the raw hardware capacity for storage and I/O is available in an infrastructure, the complexity arising from the scale and parallelism is daunting and requires significant advances in software to provide the required performance to applications.

The Storage Efficient Access (SEA) component provides the software infrastructure necessary for efficient use of the I/O hardware by applications. This is accomplished through a sequence of tightly coupled software layers, shown in Figure 8, building on top of I/O hardware at the bottom and providing application-oriented, high-level I/O interfaces at the top. Three APIs are made available for accessing SEA components: Parallel netCDF at the high-level I/O library level, ROMIO at the MPI-IO level, and Parallel Virtual File System (PVFS) at the file level.

PVFS9 can provide multiple GB/second parallel access rates, and is freely available. Above the parallel file system is software designed to aid applications in more efficiently accessing the parallel file system. Implementations of the MPI-IO interface are arguably the best example of this type of software. MPI-IO provides optimizations that help map complex data movement into efficient parallel file system operations. Our ROMIO10 MPI-IO interface implementation is freely distributed and is the most popular MPI-IO implementation for both clusters and a wide variety of vendor platforms. MPI-IO is a powerful but low-level interface that operates in terms of basic types, such as floating point numbers, stored at offsets in a file. However, some scientific applications desire more structured formats that map more closely to the application’s use, such as multidimensional datasets. NetCDF11 is a widely used API and portable file format that is popular in the climate simulation and data fusion communities. As part of the work in the SDM center, a parallel version of NetCDF (pNetCDF) was developed. It provides a new interface for accessing NetCDF data sets in parallel. This new parallel API closely mimics the original API, but is designed with scalability in mind and is implemented on top of MPI-IO. Performance evaluations using micro-benchmarks as well as application I/O kernels have shown major scalability improvements over previous efforts. Figure 9 shows schematically the concept of adding a parallel netCDF layer to eliminate serialization through a single processor.
Upcoming systems will incorporate hundreds of thousands of compute processors along with support nodes. Using POSIX and MPI-IO interfaces, I/O operations will be forwarded through a set of I/O nodes to storage targets. Work is underway to develop efficient forwarding systems to match petascale architectures and to best connect to underlying file systems, including PVFS.
Despite recent advancements in storage technologies for many data intensive applications, analysis of data remains a serious bottleneck. In traditional cluster systems, I/O-intensive tasks must be performed in the compute nodes. This produces a high volume of network traffic. One option for data analysis is to leverage resources not on the client side, but on the storage side referred to as Active Storage. The original research efforts on active storage were based on a premise that modern storage architectures might include usable processing resources at the storage controller or disk; unfortunately, commodity storage has not yet reached this point. However, parallel file systems offer a similar opportunity. Because the servers used in parallel file systems often include commodity processors similar to the ones used in compute nodes, many Giga-op/s of aggregate processing power are often available in the parallel file system. As part of the SEA layer technology, our goal in the Active Storage project is to leverage these resources for data processing. Scientific applications that rely on out-of-core computation are likely candidates for application of this technique, because their data is already being moved through the file system. The Active Storage approach allows moving computations involving data stored in a parallel file system from the compute nodes to the storage nodes. Benefits of Active Storage include low network traffic, local I/O operations, and better overall performance. The SDM center has implemented Active Storage on Lustre and PVFS parallel file systems. We plan to pursue deployment of Active Storage in biology or climate application.
