
March 2008
|
Life or Death Decision-making: The Medical Case for Large-scale, On-demand Grid Computing
|
 March 2008 |
Patient-specific medicine is the tailoring of medical treatments based on the characteristics of an individual patient. Decision support systems based on patient-specific simulation hold the potential of revolutionising the way clinicians plan courses of treatment for various conditions, such as viral infections and lung cancer, and the planning of surgical procedures, for example in the treatment of arterial abnormalities. Since patient-specific data can be used as the basis of simulation, treatments can be assessed for their effectiveness with respect to the patient in question before being administered, saving the potential expense of ineffective treatments and reducing, if not eliminating, lengthy lab procedures that typically involve animal testing.
In this article we explore the technical, clinical and policy requirements for three distinct patient-specific biomedical projects currently taking place: the patient-specific modelling of HIV/AIDS therapies, cancer therapies, and addressing neuro-pathologies in the intracranial vasculature. These patient-specific medical simulations require access to both appropriate patient data and the computational and network infrastructure on which to perform potentially very large-scale simulations. The computational resources required are supercomputers, machines with thousands of cores and large memory capacities capable of running simulations within the time frames required in a clinical setting; the validity of results not only relies on the correctness of the simulation, but on its timeliness. Existing supercomputing site policies, which institute ‘fair share’ system usage, are not suitable for medical applications as they stand. To support patient-specific medical simulations, where life and death decisions may be made, computational resource providers must give urgent priority to such jobs, and/or facilitate the advance reservation of such resources, akin to booking and prioritising pathology lab testing.
Recent advances in advance reservation and cross-site run capabilities on supercomputers mean that, for the first time, computation can be envisaged in more than a scientific research capacity so far as biomedicine is concerned. One area where this is especially true is in the clinical decision-making process; the application of large-scale computation to offer real-time support for clinical decision-making is now becoming feasible. The ability to utilise biomedical data to optimise patient-specific treatment means that, in the future, the effectiveness of a range of potential treatments may be assessed before they are actually administered, preventing the patient from experiencing unnecessary or ineffective treatments. This should provide a substantial benefit to medicine and hence to the quality of life of human beings.
Traditional medical practice requires a physician to use judgement and experience to decide on the course of treatment best suited to an individual patient’s condition. While the training and experience of physicians hone their ability to decide the most effective treatment for a particular ailment from the range available, this decision making process often does not take into account all of the data potentially available. Indeed in many cases, the sheer volume or nature of the data available makes it impossible for a human to process as part of their decision making process, and is therefore discarded. For example, in the treatment of HIV/AIDS, the complex variation inherent within data generated by analysis of viral genotype resulting in a prediction of phenotype (in terms of viral sensitivity to a number of treatments) makes the selection of treatment for a particular patient based on these predictions fairly subjective.
Patient-specific medical simulation holds the promise of evaluating tailored medical treatment based on the particular characteristics of an individual patient and/or an associated pathogen. Furthermore, approaches using simulation are based on the development of theories and models from which deductions can be made, as is the standard approach in the physical sciences and engineering. In reality, biology and medicine are still too poorly understood for deductive approaches to replace inductive ones so, in the foreseeable future, both will continue to sit side by side 1. However for clinical acceptance, verification and validation of these techniques need to be addressed. The patient-specific simulation approach contrasts with more traditional use of computer systems to support clinical decision making, such as ‘classic’ expert systems, which take a Baconian approach, allowing a clinician to infer the cause of symptoms or the efficacy of a particular treatment regime based on historical case data. An example of such a system is the MYCIN expert system 2, designed to suggest possible bacterial causes of a patient’s infection by asking a clinician a series of ‘yes’ or ‘no’ questions.
While the details vary widely between medical conditions, several basic elements are common to all fields of patient-specific medical simulation in support of clinical decision-making. Data is obtained from the patient concerned, for example from an MRI scan or genotypic assay, which is used to construct a computational model. This model is then used to perform a single simulation, or can form the basis of a complex workflow of simulations of a proposed course of treatment; for example, molecular dynamics simulations of drugs interacting with a range of viral proteins, and the results of the simulation are interpreted to assess the efficacy of treatment under consideration. The use a of simulation to assess a range of possible treatments based on data derived from the patient who is to be treated will give the physician the ability to select a treatment based on prior (simulated) knowledge of how the patient will respond to it.
The patient-specific medical simulation scenarios touched on above require access to both appropriate patient data and to the infrastructure on which to perform potentially very large numbers of complex and demanding simulations. Resource providers must furnish access to a wide range of different types of resource, typically made available through a computational grid, and to institute policies that enable the performance of patient-specific simulations on those resources. A computational grid refers to a geographically distributed collection of supercomputing resources, typically connected by high-capacity networking infrastructure, and we define grid computing as distributed computing conducted transparently across multiple administrative domains 3. For the purpose of this article, grids can also include other resources, such as medical imaging equipment and data visualisation facilities.
In order to make patient-specific simulations useful to a physician, results need to be obtained within a clinically useful timeframe, which ranges from instantaneous results to weeks, depending on the scenario. In addition to expediency of access to patient data, consideration must also be given to policy and procedures that ensure maintenance of patient confidentiality. For such an enterprise to succeed, grid computing will need to focus not only on the provision of large ‘island’ compute machines but also on the performance characteristics of the networks connecting them. The process of clinical decision making, requiring access to relevant data, timely availability of computational results, visualisation, data storage, and so on, requires infrastructure that can facilitate the transfer of gigabytes of data within clinically relevant timeframes.
When used as part of the clinical decision making process, computational resources often need to support more exotic scheduling policies than simple first come, first served, batch scheduling, which is the typical scenario seen in high-performance research computing today. Clinicians who require interactive access to machines (for example, for steering and visualisation, as well as cross-site applications, for example when performing cerebral blood flow simulations using the HemeLB code discussed later in this article) also need to be able to both schedule time on specific resources - compute and networking - and access tools to allow them to easily co-reserve those resources, so they are available when needed. This in turn leads to a demand on resource providers to implement policies and tools that allow such reservations to be made as needed and when required, so that such methodologies can be incorporated into a user's normal research activities, rather than just providing such facilities on an ad hoc basis. Moreover, the resources provided by a single grid may not always be sufficiently powerful or appropriate to run large-scale distributed models, and resources provided by multiple grids may need to be federated in order for a particular investigation to be conducted.
If these resources need to be used interactively, the problem of reservation becomes compounded since each grid has its own policies and systems for making advanced reservations, if it has any at all. Additionally, the high performance network provision between grids may also be limited or non-existent. Nevertheless, such obstacles must be overcome to make efficient use of available federated systems.
The key factor that transcends all of the current patient-specific medical simulation scenarios described in this article is the need to turn simulations around fast enough to make the result clinically relevant. This in turn means that the results can be obtained and interpreted within a timeframe on which a clinical decision is made; for example, in the HIV case described later this is roughly two weeks – the time it takes to get the results of a genotypic assay. To achieve the required turn around factor, such simulations cannot be run in a resource’s normal batch mode; they need to be given a higher priority and they require some form of on-demand computing to succeed.
We consider two different urgent computing paradigms in order to make use of supercomputing resources, provided by a grid, in clinical scenarios; the advance reservation of CPU time on a compute resource at some specific point in the future, and the pre-emption of running jobs on a machine by some ‘higher priority’ work. The two paradigms apply to slightly different situations; the former would be of most use when a clinician knows in advance that a simulation needs to be performed at a specific time, for example an interactive brain blood-flow simulation run for a surgeon while planning or conducting a surgical procedure. The second paradigm is most useful when a medical simulation needs to be performed urgently, but the need for the simulation is not known in advance. An example of this latter simulation would be where a clinician encounters a HIV patient and urgently needs to compute the efficacy of a series of inhibitor drugs in relation to the patient’s specific HIV mutation.
There is crossover between the two different urgent computing paradigms considered, and to a certain extent both apply to each of the situations mentioned. We favour a combination of both paradigms, to give clinicians and scientists the greatest amount of flexibility possible for the work they need to conduct. We discuss the technical aspects of the two paradigms in greater detail below.
Several systems exist to allow users to easily co-reserve time on grid resources. GUR (Grid Universal Remote)4 is one such system, developed at San Diego Supercomputer Center (SDSC). The GUR tool is a python script, which builds on the ssh and scp commands to give users the ability to make reservations of compute time and co-schedule jobs. GUR is installed on the SDSC, National Center for Supercomputing Applications (NCSA) and Argonne National Laboratory (ANL) TeraGrid IA-64 systems, and is expected to be available at other TeraGrid sites soon.
HARC (Highly Available Robust Co-scheduler) is one of the most robust and widely deployed open-source systems that allows users to reserve multiple distributed resources in a single step 5. These resources can be of different types, including multiprocessor machines and visualisation engines, dedicated network connections, storage, the use of a scientific or clinical instrument, and so on. HARC can be used to co-allocate resources for use at the same time, for example, within a scenario in which a clinical instrument is transferring data over a high-speed network link to remote computational resources for real-time processing. It can also be used to reserve resources at different times for the scheduling of workflow applications. We envisage clinical scenarios within which patient-specific simulations can be timetabled and reserved in advance, via the booking of an instrument, the reservation of network links and storage facilities, followed by high-end compute resources to process data, and finally the use of visualisation facilities to interpret the data for critical clinical decisions to be made.
Currently, HARC can be used to book computing resources and lightpaths across networks based on GMPLS (Generalised Multi-protocol Label Switching) with simple topologies. HARC is also designed to be extensible, so new types of resources can be easily added; it is this that differentiates HARC from other co-allocation solutions. There are multiple deployments of HARC in use today: the US TeraGrid, the EnLIGHTened testbed in the United States, the regional North-West Grid in England, and the National Grid Service (NGS) in the UK. We use HARC on a regular basis to make single and multiple machine reservations, within which we are able to run numerous applications including HemeLB (see Section 4.1).
SPRUCE (SPecial PRiority and Urgent Computing Environment) 6 is an urgent computing solution that has been developed to address the growing number of problem domains where critical decisions must be made quickly with the aid of large-scale computation. SPRUCE uses simple authentication mechanisms, by means of transferable ‘right of way’ tokens. These tokens allow privileged users to invoke an urgent computing session on pre-defined resources, during which time they can request an elevated priority for jobs. The computations can be run at different levels of urgency; for example, they can have a ‘next to run’ priority, such that the computation is run once the current job on the machine completes, or ‘run immediately,’ such that existing jobs on the system are removed, making way for ‘emergency’ computation in a pre-emptive fashion, the most extreme form of urgent computing. The neurovascular blood-flow simulator, HemeLB (discussed in Section 4.1) has been used with SPRUCE in a ‘next to run’ fashion on the large scale Lonestar cluster at the Texas Advanced Computing Center (TACC), and was demonstrated live on the show floor at SuperComputing 2007, where real-time visualisation and steering were used to control HemeLB within an urgent computing session.
The TeraGrid also provides a contrasting solution to the need to run urgent simulations on its resources. SDSC provide an ‘On-Demand’ computer cluster, made available to researchers via the TeraGrid, to support scientists who need to make use of urgent scientific applications. The cluster is configured to give top priority to urgent simulations, where results of the simulation are needed to plan responses to real-time events. When the system is not being used for on-demand work, it runs normal batch compute jobs, similar to the majority of other TeraGrid resources. Many of the current urgent scenarios considered cover the need to anticipate the effects of natural disasters, such as earthquakes and hurricanes, by performing simulations to predict possible consequences while the event is actually happening. Patient-specific medical simulations present another natural set of use cases for the resource.
In this section, we discuss three examples of patient-specific medicine where computational approaches are showing promise. Although the overall pathologies in each case are similar from patient to patient, the underlying details of the pathology can differ dramatically. In the case of HIV/AIDS and cancer treatments, the underlying mutations of these conditions are related to the genotype of the patient, and in the case of neurovascaular pathologies, the cerebral vascular structure differs considerably between individuals, so that each person will exhibit different blood flow dynamics. All these cases exemplify ‘patient-specific’ approaches, since the treatment is based on genotypic and/or phenotypic information obtained from the patient.
Cardiovascular disease is the cause of a large number of deaths in the developed world 7. Cerebral blood flow behaviour plays a crucial role in the understanding, diagnosis and treatment of this disease. The problems are often due to anomalous blood flow behaviour in the neighbourhood of bifurcations and aneurysms within the brain; however, the details are not very well understood.
Experimental studies are frequently impractical owing to the difficulty of measuring flow behaviour in humans; however, X-ray and magnetic resonance imaging angiography (MRA) enable non-invasive static and dynamical data acquisition 8. Indeed, some studies have revealed relationships between specific flow patterns around walls and cardiovascular diseases such as atherosclerosis 9.
Today, such imaging methods represent a very important tool for diagnosis of various cardiovascular diseases, together with the design of cardiovascular reconstructions and devices for the enhancement of blood flow. Notwithstanding these advances in measurement methods, modelling and simulation undoubtedly have a crucial role to play in haemodynamics. Simulation, for example, furnishes the clinician with the possibility of performing non-invasive virtual experiments to plan and study the effects of certain courses of (surgical) treatment with no danger to the patient, offering support for diagnosis, therapy and planning of vascular treatment 10. Modelling and simulation also offer the prospect of providing clinicians with virtual patient-specific analysis and treatments.
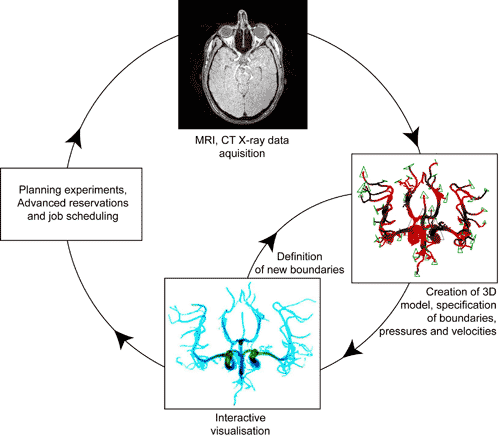
Reaching the goal of blood flow modelling and simulation is dependent on the availability of computational models of sufficient complexity and power. Furthermore, the neurovascular system varies between every single person, and so any computational approach will require patient-specific data. The aforementioned imaging techniques are used to provide data for such simulations. Furthermore, the computational fluid ‘solver’ used must itself be numerically highly efficient and provide scientists and neurosurgeons with the ability to manipulate and visualise the associated large data sets. The intricate geometry of the fluid vessels and treatment of fluid boundary conditions at such walls are also very difficult for traditional continuum fluid dynamics models to handle. Instead, a lattice-Boltzmann (LB) method, coined HemeLB, offers an attractive alternative. A major feature of HemeLB is real-time rendering and computation; fluid flow data is rendered in-situ on the same processors as the LB code, and sent, in real-time, to a lightweight client on a clinical workstation (Figure 1). The client is also used to steer the computation in real time, allowing the adjustment of physical parameters of the neurovascular system, along with visualisation-specific parameters associated with volume rendering, isosurface rendering, and streamline visualisation.

HemeLB is intended to yield patient-specific information, which helps plan embolisation of arterio-venous malformations and aneurysms, amongst other neuro-pathologies. Using this methodology, patient-specific models can be used to address issues with pulsatile blood flow, phase differences and the effects of treatment, all of which are potentially very powerful both in terms of understanding neurovascular patho-physiology and in planning patient treatment.
The software environment used in this project aims to bring to the forefront details and processes clinicians need to be aware of, such as (i) the process of image segmentation to obtain a 3D neurovascular model, (ii) the specification of pressure and velocity boundary conditions, and (iii) the real-time rendered image (Figure 2). Clinicians are not concerned with where simulations are running, nor the details of reservations, thus features such as advanced reservations and emergency computing capabilities, job launching and research selection are all done behind the scenes. This environment is particularly important given the time scales involved in the clinical decision making process in the treatment of aterio-venous malformations and aneurysms. From the acquisition of a 3D dataset (which is typically 2 to 4 GB in size), to the next embolisation, a time scale of 15 to 20 minutes is typical, and for such computational approaches to be clinically relevant, we have to fit into this time scale. There are also preventative scenarios that can be envisioned; patients could be subjected to such simulations in advance of vascular pathologies developing, averting future problems with interventional treatments.
A major problem in the treatment of AIDS is the development of drug resistance by the human immuno-deficiency virus (HIV). HIV-1 protease is the enzyme that is crucial to the role of the maturation of the virus and is therefore an attractive target for HIV/AIDS therapy. Although several effective treatment regimes have been devised which involve inhibitors that target several viral proteins 11, the emergence of drug resistant mutations in these proteins is a contributing factor to the eventual failure of treatment.
Doctors have limited ways of matching a drug to the unique profile of the virus as it mutates in each patient. A drug treatment regimen is prescribed using knowledge-based clinical decision support software, which attempts to determine optimal inhibitors using existing clinical records of treatment response to various mutational strains. The patient’s immune response is used as a gauge of the drug's effectiveness and is periodically monitored so that ineffective treatment can be minimised through an appropriate change in the regimen. The FP6 EU project ‘Virolab’ is attempting to enhance the efficacy of clinical decision support software, through a unification of existing databases, as well as integration with means of assessing drug resistance at the molecular level 12.
At the molecular level, it is the biochemical binding affinity (free energy) with which an inhibitor binds to a protein target that determines its efficacy. Experimental methods for determining biomolecular binding affinities are well established and have been implemented to study the in-vitro resistance conferred by particular mutations. These in turn add invaluable information to any decision support system, but are limited as studies are performed usually on key characteristic mutations and not with respect to the unique viral sequence of a patient. An exhaustive experimental determination of drug binding affinities in a patient-specific approach is far too costly and time-consuming to perform in any clinically relevant way.

Computational methods also exist for determining biomolecular binding affinities. In a recent study 13, the effectiveness of the drug saquinavir was tested against the wildtype HIV-1 protease, along with three drug-resistant strains using free energy methods in molecular dynamics (MD) simulations (Figure 3). The protocol implemented by the study gave accurate correlations to similar experimentally determined binding affinities. Furthermore, the study made use of a tool, the Binding Affinity Calculator (BAC), for the rapid and automated construction, deployment, implementation and post processing stages of the molecular simulations across multiple supercomputing, grid-based resources. The BAC is built on top of the Application Hosting Environment (AHE) 14, a web services environment designed to hide the complexity of application launching from the scientific end user of the grid. The AHE makes use of Globus Toolkit versions 2 and/or 4 for job submission, and GridFTP for data transfer between resources.
BAC automates binding affinity calculations for all nine drugs currently available to inhibit HIV-1 protease and for an arbitrary number of mutations away from a given wildtype sequence. Although the applicability of the method in the saquinavir-based study still needs to be established for all other inhibitors, the scope of BAC is enormous as it offers an automated in-silico method for assessing the drug resistance for any given viral strain. The turn around time using BAC for such studies is seven days (per drug/protease system) with optimal computational resources; this is more than suitable for the timescales required for effective clinical decision support. Given enough computational power such that binding affinity calculations can be routinely applied, the potential to achieve patient-specific HIV decision support may then become realistic.
The identification and treatment of cancer exists on various levels, from the large-scale view of tumour growth down to individual molecular interactions. The treatment of cancer often takes two directions, using targeted radiotherapy to kill malignant cells, while also using tumour-growth inhibitors in an attempt to selectively target and kill tumourous cells. The effectiveness of particular chemotheraputic treatment differs from patient to patient, with some courses of treatment not being effective at all.
A new generation of anticancer drugs are part of an approved scheme called ‘targeted therapy,’ in which anticancer drugs are directed against cancer-specific molecules and signalling pathways. These are designed to interfere with a specific molecular target, usually a protein that plays a crucial role in tumour cell growth and proliferation. Receptor tyrosine kinases (RTKs) are an example; they are cell surface proteins that can be used as targets to control tumour growth in various preclinical treatment models. Tyrosine kinase inhibitors (TKI) interfere with the related cell signalling pathways and thus allow target-specific therapy for selected malignancies. In fact, some TKIs have been approved for use in cancer therapy, and others are in various stages of clinical trials.
RTKs have been found to be over-expressed or mutated in tumour cells, and these mutations allow cancer cells to develop drug resistance. Clinical studies have shown a strong correlation between a reduction in the response to treatment with TKIs and the presence of these mutations, where the resistance is introduced by preventing or weakening the binding of the receptor to the targeted TKI.
The binding of the tumour-growth inhibitors to cell receptors is identical to small molecule-protein or protein-protein interactions. Molecular dynamics techniques can be used to study these interactions in atomistic detail, and to predict the effect of different receptors and mutations on inhibitor binding affinities. Using patient-specific data, such as the RTK mutation, which is expressed on tumourous cells, MD techniques can be used to rank the binding affinities, and therefore the effectiveness of various treatments against a patient-specific case.
Using a grid-infrastructure, turnaround times can be dramatically accelerated. MD simulations, particularly for the case of various inhibitors and possibly various targets, can be independently run by being farmed off to various grid resources. Providing turnaround times of five days will ensure that the findings are clinically relevant and become part of the clinical decision making process. One of the aims within this project is to develop a work-flow tool, which will use the AHE to permit the automated running of such patient-specific simulations, hiding the unnecessary grid details from clinicians.
Patient-specific medical simulation holds the promise of revolutionising the diagnosis and treatment of many different medical conditions, by making use of advanced simulation techniques and high performance compute resources. For computational medicine to be of use in modern clinical settings, the timeliness with which results are delivered is of primary concern. Results need to be generated in a timeframe that is useful to the clinician initiating the simulation results; that is, they must be generated in time to inform the treatment regime or procedure under consideration. In the case of neurosurgical treatments, this is in the order of 15 to 20 minutes. In the case of HIV or cancer pathology reports, this is in the order of 24 to 48 hours.
Due to the urgent requirements of patient-specific simulations, the current standard model of high performance compute provision, the batch queue model, is of no use. Simulations have to fit into existing clinical processes; clinical processes cannot be altered to adapt to a batch compute model, as very often a simulation will be used to inform an urgent life or death decision. Because of this, technologies that enable and facilitate urgent computing are of great relevance to the emerging field of patient-specific simulation. Advance reservation tools such as HARC and urgent computing systems such as SPRUCE are essential for making patient-specific medical simulation a reality when using general purpose, high performance compute resources that typically run a wide range of different tasks.
Systems such as HARC and SPRUCE were initially conceived to support the submission of very infrequent on-demand jobs, for example climate models, that typically are only run in emergency situations, such when a hurricane is looming; running simulations at this frequency on general purpose compute resources, such as those available on the TeraGrid, have a negligible effect on the users of the system as a whole. In the case of an urgent simulation using SPRUCE, a limited set of users who had their jobs pre-empted would not notice anything different. Patient-specific medical simulations are of a different nature; a successful patient-specific simulation technique will likely have thousands, or even tens of thousands, of possible patients that it could be performed for. The possible level of compute time required will dwarf the current urgent-computing policies and resources in place.
Patient-specific medical simulation raises several moral, ethical and policy questions that need to be answered before the methodologies can be put to widespread use. Firstly there is the question of the availability of resources to perform such simulations. The compute power currently made available through general purpose scientific grids, such as the TeraGrid or UK NGS, is not enough to satisfy the potential demand of medical simulation. The scarcity of resources raises the question of how such resources will be allocated. Which patients will benefit from medical simulations? Will it be based on the ability to pay? Secondly there is the question of data privacy. Sensitive clinical information is often kept on highly secure hospital networks, and the owners and administrators of such networks are often loath to let any data move from it onto networks over which they have no control, which is necessary if the data is to be shipped to a remote site and used in a simulation. Using such data on ‘public’ grid resources requires it to be suitably anonymised, so that even if it were to fall into the wrong hands it could not be traced back to the patient it was taken from.
We believe that as such tasks become more widespread and embedded in the clinical process, the market will start to address the first question raised above. Already, many companies are starting to provide utility compute services, such as Amazon’s Elastic Compute Cloud 15, which allows the public to purchase computational cycles. If a market was created for running medical simulations on demand, then we believe it likely that utility compute providers will move to supply the necessary compute services. Although it is uncertain how a pricing model will work in reality, it is likely that the utility compute model will drive down the costs of such simulations, and where the performance of simulations is shown to make a treatment regime more efficient, it is likely that the cost could be met from the money saved. The second question needs to be addressed by medical data managers and government regulators. Once enabling policies have been developed, the process of routinely anonymising data and shipping it from a hospital network or storage facility will become routine. Such a system of anonymisation is being implemented in the neurovascular project discussed, involving discussions with technical network administrators and management from the UK National Health Service (NHS).
It is essential that a dialogue is joined between governments, researchers, health professionals and business into how the infrastructure needed to perform patient specific medical simulations can be performed on a routine basis. The benefits of performing such simulations are too great to be ignored and, in addition to the case studies presented, we believe that computational simulation will be used in more and more medical scenarios. In the vision that patient-specific medical simulations become a day to day reality in the treatment of patients, vast quantities of simulation data will be available alongside traditional medical data. With parallel advances in data warehousing, data-mining and computational grids, the enhancement of medical practice using simulation will one day become a reality.