Opal is developed by NBCR as a Java based toolkit that automatically wraps any legacy applications with a Web services layer that is fully integrated with Grid Security Infrastructure (GSI) based security, cluster support, and data management.32 51 The advantage of using OPAL is that the application may be launched using any Web service client, because the WSDL defines using the standard protocol for how the service may be accessed, and it provides the basic HTTP access to results, as well as any metadata that describes how the results may be handled (Figure 6A). This ‘deploy once use by many’ feature of a Web service is a key ingredient for achieving interoperability. Because OPAL manages the interaction with the grid architecture, the grid is transparent to the user. In addition, workflow tools like Kepler52 may easily compose web services based workflows through a common interface (Figure 6B).
GAMA, co-developed by NBCR,53 is a GSI-based security service that manages X.509 user credentials on behalf of users and supports SOAP-based applications (Figure 7A). The server component leverages existing software packages such as CAS, MyProxy, and CACL with the GAMA version 2 supporting other CA packages such as NAREGI. A portlet component provides the administrative interface to the server. As security is a sensitive and critical issue in the production use of the cyberinfrastructure, GAMA allows any (Figure 7) organization to create their own certificate authority, manage their user certificates, secure the SOAP communications using HTTPS and mutual authentication, and integrate seamlessly with portals and rich clients. For example, GAMA is used by GridSphere based portals,54 Gemstone55 and PMV/Vision.
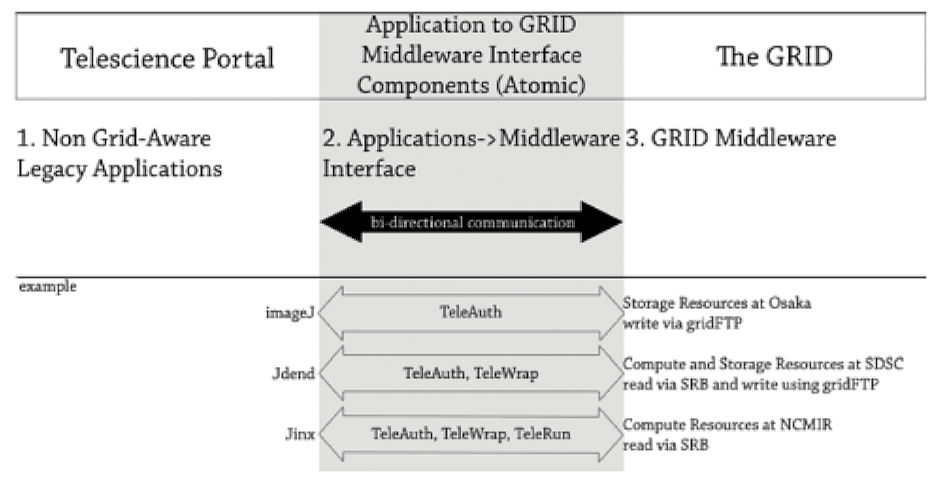
The ATOMIC bundle (Figure 7B), developed for the Telescience Project by NBCR, provides a fabric for seamless interoperability among user interfaces (web portals and applications) and externally addressable grid resources (instruments and computers) inside a grid portal environment.56 There is a unified programming API for basic scientific research, which liberates the users and programmers from having to learn about the grid. TeleAuth is a specialized version of GAMA. TeleWrap provides seamless access to remote data transparently and has been used successfully in the Telescience portal and EMWorkspace. TeleRun provides a higher level of abstraction than Opal-provided services and may further shield developers from the details of grid execution environment. ATOMIC also provides persistent session information during a particular Telescience session, so that all user portlets have access to the same session information regardless of the specific portlet being used.